Archive: Microbial Biogeography Within Individual Trees
On a gray morning in Boston, Massachusetts, I met with Ned Friedman, the director of the Arnold Arboretum, and Peter Del Tredici, a senior scientist at the Arboretum and ginkgo guru. We were all excited to begin work sampling the microbiota of a few ginkgo trees. We were also excited to use the arboretum's cherry picker, a finely tuned piece of equipment capable of hoisting researchers 15+ meters in the air.
Although the execution was anything but simple, the idea behind the project was relatively straightforward. We sought to answer a couple basic research questions fundamental to the field of plant-microbe interactions:
To what extent does bacterial diversity and community composition vary within an individual tree?
Can this variability be predicted based on factors such as tissue type and location within the tree?
With these questions in mind, we headed to the trees. We planned to sample three of them. They were all planted at the same time and were relatively similar, although they varied somewhat in height. Our plan was to take samples from relatively consistent locations in each tree. This way, we would have, in essence, three replicates for each sampling location. For instance, we would have a 'south side-third branch up-5 growth scars out' sample for each tree. I should also mention that one reason we sampled the ginkgo trees is because of their remarkably consistent morphology that made it relatively easy to know how old specific branch segments were (we just counted back the number of growth scars that are formed every year on each branch). This was particularly helpful when comparing sampling locations across the three trees.
After surveying the trees and deciding on a plan of action, we were almost ready to proceed. I placed a quick call to my advisor, Noah Fierer, to make sure he agreed with our plan, and everything was in place. We ambitiously planned to collect more than one hundred samples per tree -- no small feat given that each sample had to be carefully secured without contaminating it with microbes from my body, other samples, or our tools. And, many of these samples had to be collected from the awkwardly sized cherry picker bucket (see photo to left).
This is where I could say that everything went wrong, but it didn't. The weather even cleared, which made for a pleasant, albeit tedious experience. Lucky for me, my girlfriend, Samantha Weintraub, agreed to help with the sample collection and data recording. Even more fortunate was that she had way more field experience than me through work on her own research -- This made her critical for keeping all the samples organized and appropriately labeled.
In the end, we had roughly 315 bark swabs and leaf samples collected and ready for bacterial analysis. To analyze the bacterial communities on each sample, we used a high throughput sequencing approach described here.
Spatial Variation in Diversity
One way to coarsely address question 1 was to see whether different tissue types within the same tree harbor consistently different microbial communities. We sampled four different types of tissues (at least we put them in four categories): trunk, branch, newly grown branches, and leaves. First, we looked at species diversity -- the number of unique species within a given sample. Did this differ across the multiple tissue types?
In fact, there were differences. Trunk samples were more than twice as diverse (i.e. had twice the number of unique species) as young tissues (leaves and newly grown branch samples). This finding is interesting in that it suggests there is a narrower set of early colonizing bacteria on young tissues, and communities become more diverse as they age.
We also looked to see whether diversity was variable within a particular tissue type. For instance, we found that bark tissues had more diverse bacterial communities in the interior of the tree and less diverse communities on their periphery. The consistency of this pattern was striking.
Spatial Variation in Community Composition
Next, we wanted to address whether the bacterial community composition differed across tissue types or spatially within the individual trees. We compared whole communities by plotting them in an ordination - a technique that represents communities as a point in two dimensional space. The graph on the right shows these broad scale differences: Young tissues (green branches and leaves) tended to have similar communities, while bark tissues had more dissimilar communities.
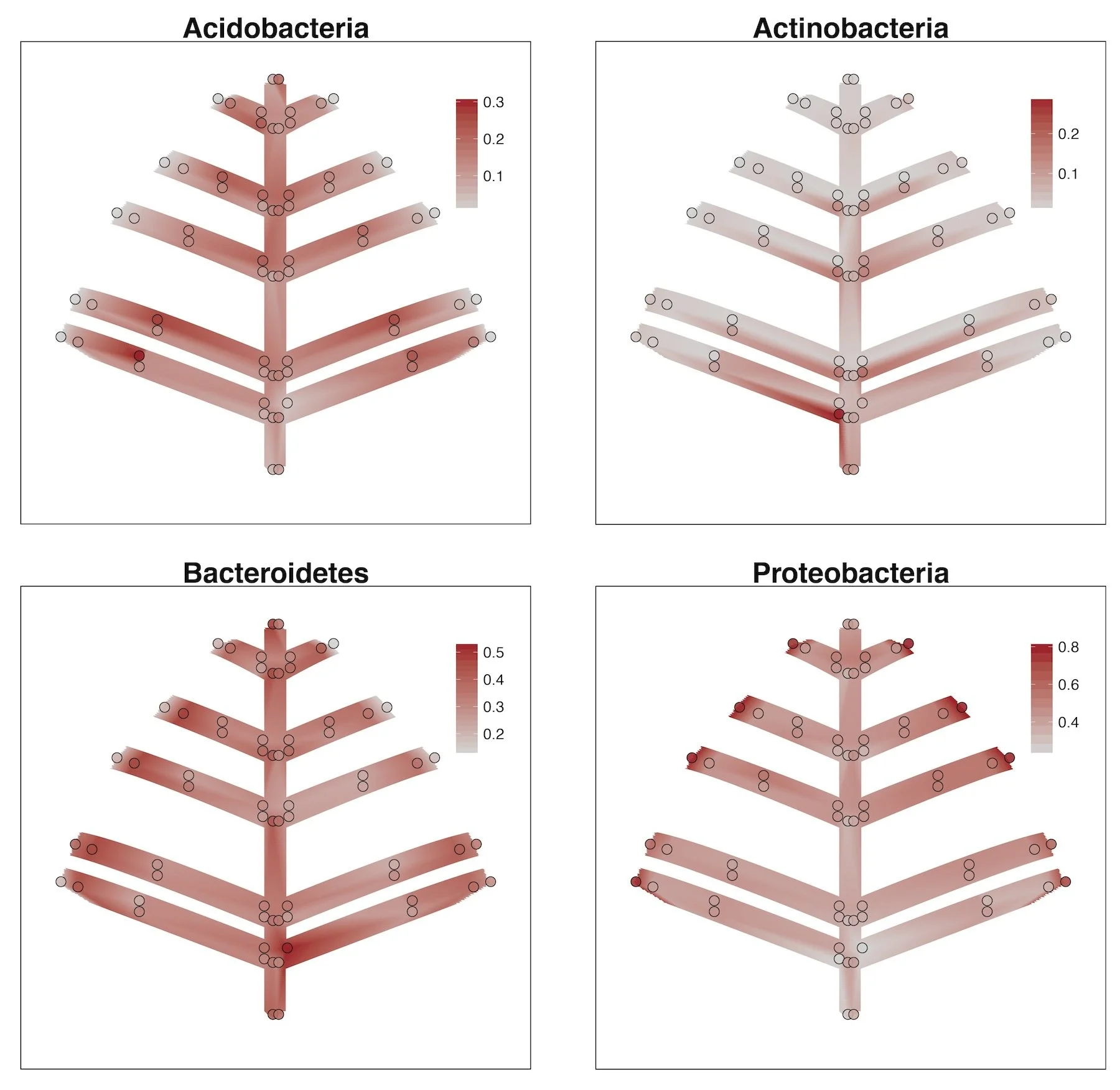
As with diversity, we also looked at variation in community composition within the individual trees. The figure to the left shows the spatial variation of the four most abundant bacterial taxa across the bark samples (click for an enlarged image). The figure shows distinct patterns for each of these groups - Acidobacteria tend to be relatively more abundant on the interior samples, while Proteobacteria tended to be relatively more abundant on the more peripheral samples. These intriguing patterns beg further hypotheses. For example, do these bacterial groups show better or worse due to intrinsic lifestyles or capabilities?
Meaning Behind the Results
There are a large number of studies investigating interactions between plants and microbes, but often times, only single samples from the plants are used to represent an individual plant. The findings from this study show that using this approach can be misleading - a sample from one part of the plant could likely have a very different bacterial community from another part.
After I collected the last sample, I asked to be hoisted as high as possible in the cherry picker. Towering 10 meters above the canopy of the ginkgo, I looked around and noticed the diverse array of other trees that went unnoticed while focusing on the ginkgo. I wondered if this was appropriate symbolism for researchers' previous focus on culturable microbes and our current efforts to uncover the meaning behind microbial communities as a whole. Perhaps not, but this study does help reinforce the idea that looking in new places solicits new questions.
More Info
Peer-reviewed journal article: link
Contact: Jon Leff
Originally posted: 11/18/2014